-Search query
-Search result
Showing 1 - 50 of 1,124 items for (author: cai & s)

EMDB-36779: 
The Anoxybacillus pushchinoensis ORF-less Group IIC Intron HYER1 with 10-nt TRS at symmetric apo state
Method: single particle / : Zhu HZ, Liu JJG

EMDB-40248: 
CRISPR-Cas type III-D effector complex
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40250: 
CRISPR-Cas type III-D effector complex bound to a self-target RNA in the pre-cleavage state
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40251: 
CRISPR-Cas type III-D effector complex bound to self-target RNA in a post-cleavage state
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40276: 
CRISPR-Cas type III-D effector complex consensus map
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40296: 
CRISPR-Cas type III-D effector complex local refinement map
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40297: 
CRISPR-Cas type III-D effector complex bound to a target RNA local refinement map
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40298: 
CRISPR-Cas type III-D effector complex bound to a target RNA consensus map
Method: single particle / : Schwartz EA, Taylor DW

PDB-8s9t: 
CRISPR-Cas type III-D effector complex
Method: single particle / : Schwartz EA, Taylor DW

PDB-8s9v: 
CRISPR-Cas type III-D effector complex bound to a self-target RNA in the pre-cleavage state
Method: single particle / : Schwartz EA, Taylor DW

PDB-8s9x: 
CRISPR-Cas type III-D effector complex bound to self-target RNA in a post-cleavage state
Method: single particle / : Schwartz EA, Taylor DW

EMDB-37985: 
Cryo-EM structure of adenosine receptor A3AR bound to CF101
Method: single particle / : Cai H, Xu Y, Xu HE

EMDB-37986: 
Cryo-EM structure of adenosine receptor A3AR bound to CF102
Method: single particle / : Cai H, Xu Y, Xu HE
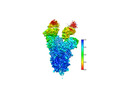
EMDB-37240: 
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Method: single particle / : Cao L, Wang X

EMDB-37241: 
The interface structure of Omicron RBD binding to 5817 Fab
Method: single particle / : Cao L, Wang X

PDB-8khc: 
SARS-CoV-2 Omicron spike in complex with 5817 Fab
Method: single particle / : Cao L, Wang X

PDB-8khd: 
The interface structure of Omicron RBD binding to 5817 Fab
Method: single particle / : Cao L, Wang X

EMDB-35931: 
cryo-EM structures of Ufd4 in complex with Ubc4-Ub
Method: single particle / : Ai HS, Mao JX, Wu XW, Cai HY, Pan M, Liu L

PDB-8j1r: 
cryo-EM structures of Ufd4 in complex with Ubc4-Ub
Method: single particle / : Ai HS, Mao JX, Wu XW, Cai HY, Pan M, Liu L

EMDB-38795: 
Cryo-EM structure of the [Pyr1]-apelin-13-bound human APLNR-Gi complex
Method: single particle / : Wang W, Ji S, Zhang Y

PDB-8xzg: 
Cryo-EM structure of the [Pyr1]-apelin-13-bound human APLNR-Gi complex
Method: single particle / : Wang W, Ji S, Zhang Y

EMDB-38794: 
Cryo-EM structure of the WN561-bound human APLNR-Gi complex
Method: single particle / : Wang W, Ji S, Zhang Y

EMDB-38796: 
Cryo-EM structure of the MM07-bound human APLNR-Gi complex
Method: single particle / : Wang W, Ji S, Zhang Y

EMDB-38797: 
Cryo-EM structure of the CMF-019-bound human APLNR-Gi complex
Method: single particle / : Wang W, Ji S, Zhang Y

EMDB-38798: 
Cryo-EM structure of the WN353-bound human APLNR-Gi complex
Method: single particle / : Wang W, Ji S, Zhang Y

PDB-8xzf: 
Cryo-EM structure of the WN561-bound human APLNR-Gi complex
Method: single particle / : Wang W, Ji S, Zhang Y

PDB-8xzh: 
Cryo-EM structure of the MM07-bound human APLNR-Gi complex
Method: single particle / : Wang W, Ji S, Zhang Y

PDB-8xzi: 
Cryo-EM structure of the CMF-019-bound human APLNR-Gi complex
Method: single particle / : Wang W, Ji S, Zhang Y

PDB-8xzj: 
Cryo-EM structure of the WN353-bound human APLNR-Gi complex
Method: single particle / : Wang W, Ji S, Zhang Y

EMDB-35449: 
Eaf3 CHD domain bound to the nucleosome
Method: single particle / : Zhang Y, Gang C

EMDB-35450: 
Cryo-EM structure of the Rpd3S core complex
Method: single particle / : Zhang Y, Gang C

EMDB-35455: 
Rpd3S bound to the nucleosome
Method: single particle / : Zhang Y, Gang C

EMDB-37849: 
Cryo-EM structure of CB1-beta-arrestin1 complex
Method: single particle / : Liao Y, Zhang H, Shen Q, Cai C

PDB-8wu1: 
Cryo-EM structure of CB1-beta-arrestin1 complex
Method: single particle / : Liao Y, Zhang H, Shen Q, Cai C

EMDB-37390: 
Cryo-EM structure of peptide free and Gs-coupled GIPR
Method: single particle / : Cong ZT, Zhao FH, Li Y, Luo G, Zhou QT, Yang DH, Wang MW, Dai AT, Shen DD, Zhang Y, Xia T, Stevens RC, Xu HE, Zhao LH

EMDB-37504: 
Cryo-EM structures of peptide free and Gs-coupled GLP-1R
Method: single particle / : Cong ZT, Zhao FH, Li Y, Luo G, Zhou QT, Yang DH, Wang MW
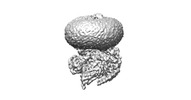
EMDB-37505: 
Cryo-EM structures of peptide free and Gs-coupled GCGR
Method: single particle / : Cong ZT, Zhao FH, Li Y, Luo G, Zhou QT, Yang DH, Wang MW

PDB-8wa3: 
Cryo-EM structure of peptide free and Gs-coupled GIPR
Method: single particle / : Cong ZT, Zhao FH, Li Y, Luo G, Zhou QT, Yang DH, Wang MW

PDB-8wg7: 
Cryo-EM structures of peptide free and Gs-coupled GLP-1R
Method: single particle / : Cong ZT, Zhao FH, Li Y, Luo G, Zhou QT, Yang DH, Wang MW

PDB-8wg8: 
Cryo-EM structures of peptide free and Gs-coupled GCGR
Method: single particle / : Cong ZT, Zhao FH, Li Y, Luo G, Zhou QT, Yang DH, Wang MW

EMDB-37516: 
BA.2(S375) Spike (S6P)/hACE2 complex
Method: single particle / : Wei X, Zhang Z

EMDB-37517: 
Local refinement of RBD-ACE2
Method: single particle / : Wei X, Zhang Z

EMDB-36678: 
PSI-AcpPCI supercomplex from Amphidinium carterae
Method: single particle / : Li ZH, Li XY, Wang WD

EMDB-36742: 
PSI-AcpPCI supercomplex from Symbiodinium
Method: single particle / : Li ZH, Li XY, Wang WD

EMDB-36743: 
PSI-AcpPCI supercomplex from Symbiodinium
Method: single particle / : Li XY, Li ZH, Wang WD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search








 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
